Bioinformatics Concepts
Background
CosmosID provides a platform to upload, process, and manage your metagenomic samples. To help you understand how our bioinformatics analysis works we will define a few terms.
kmer - a kmer is a nucleotide sequence of a certain length. It is common in genomics to select all possible kmers of a fixed length for each read in a sample, for example.
wgs - whole genome shotgun sequencing - with this method of DNA sequencing, all microbial DNA in the sample is fragmented into small pieces for next-generation sequencing.
shotgun metagenomics - using wgs sequencing as described above, the CosmosID algorithms identify microorganisms based on the entire genomes of the organisms that are in our database.
amplicon/16S/ITS - unlike shotgun metagenomics, amplicon (or 16S/ITS) analysis looks only at the relevant conserved ribosomal RNA gene or genes, not the entire genome for identification.
CosmosID Curated Databases and Patented Algorithm Info
The CosmosID databases are organized phylogenetically and contain hundreds of millions of biomarker/n-mer sequences. The markers represent both coding and non-coding sequences uniquely identified by taxon and/or distinct nodes of phylogenetic trees. This means that the tree structure was created based on genomic relatedness of organisms rather than predetermined taxonomy based on phenotype. This allows CosmosID to have a high degree of accuracy in identifying microorganisms based on their DNA in metagenomic samples. It also helps identify the closest match to genomes that do not have strain level references in the database (if, for example, they have never been sequenced before). The reference database constitutes both publicly available genomes or gene sequences through NCBI- RefSeq/WGS/SRA/nr, PATRIC, M5NR, IMG, ENA, DDBJ, CARD, ResFinder, ARDB, ARG-ANNOT, mvirdb, VFDB etc., as well as a subset of genomes sequenced by CosmosID and its collaborators.
The Algorithm has two separable comparators: the first consists of a pre-computation phase for the reference database and a per-sample computation. The input to the pre-computation phase is a reference microbial genome or antibiotic resistance and virulence gene database, and its output is a phylogeny tree, together with sets of variable length k-mer fingerprints (biomarkers) that are uniquely identified with distinct branches and leaves of the tree. The second per-sample, computational phase searches the hundreds of millions of short sequences or contigs from draft assembly against the k-mer fingerprint sets and looks for exact matches. The resulting statistics are analyzed to give fine-grain compositional statistics and relative abundance estimates
Types of Databases
Organism databases:
- Bacteria
- Viruses
- Fungi
- Protists
- Respiratory Viruses
Gene databases:
- Antibiotic Resistance
- Virulence Factor
Identification at Different Taxonomic Levels
Figure 1: ID and abundance at each taxonomic level
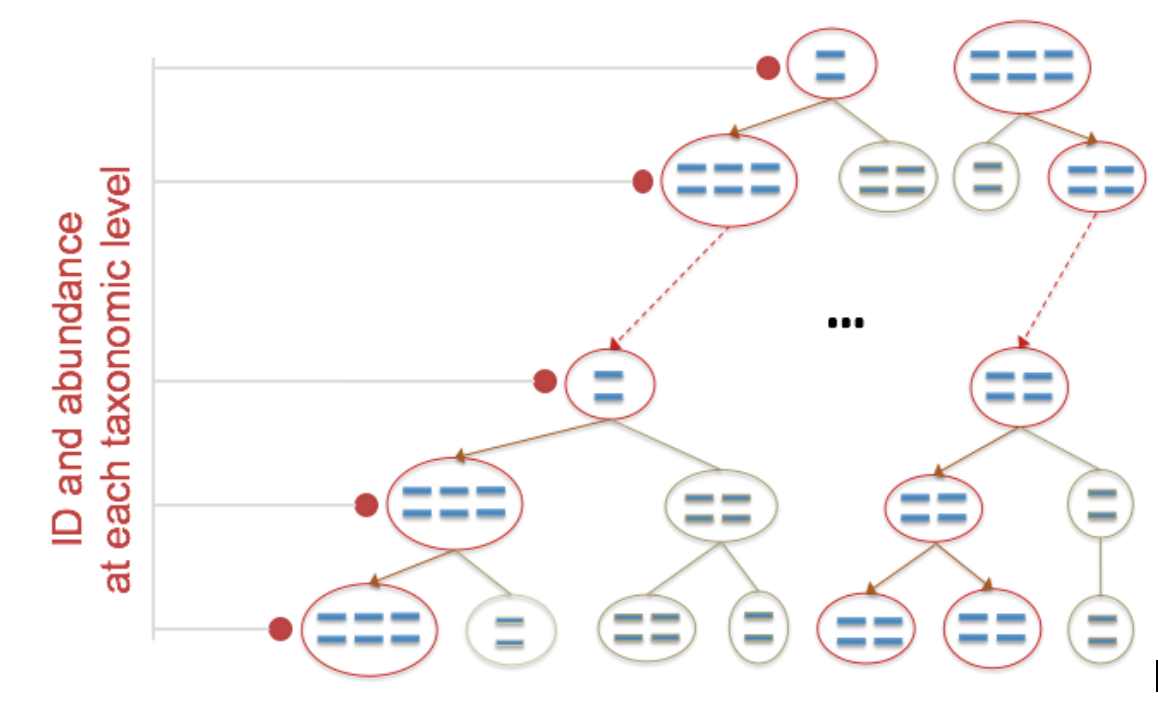
In Figure 1 you can see how kmers are mapped to taxonomic levels. Kmers are identified that are unique to each reference in the CosmosID database. Identification is made at the lowest taxonomic level possible, depending on which kmers are found in the sequenced sample.
Updated about 1 year ago
