How to Upload your Zymo Standard
To navigate to the Microbiome Standard page, click the Upload Microbiome Standard icon
Sequence QC
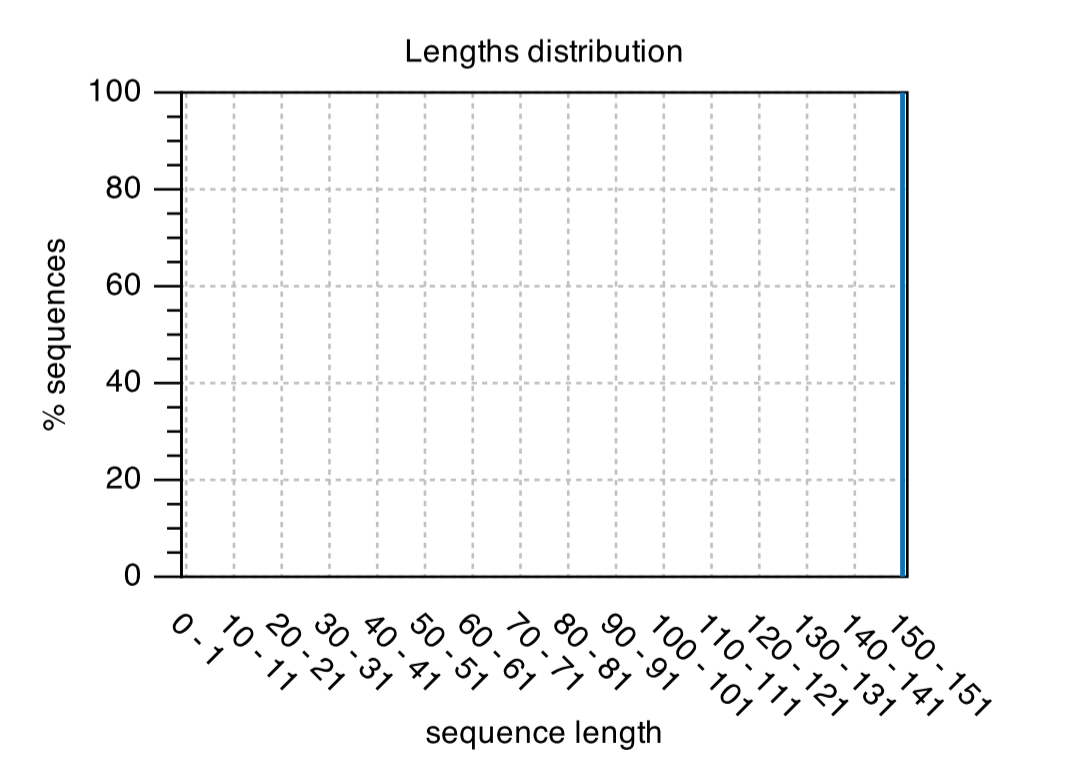
After running your microbiome standard you will see a QC report for your sample.Length Distribution
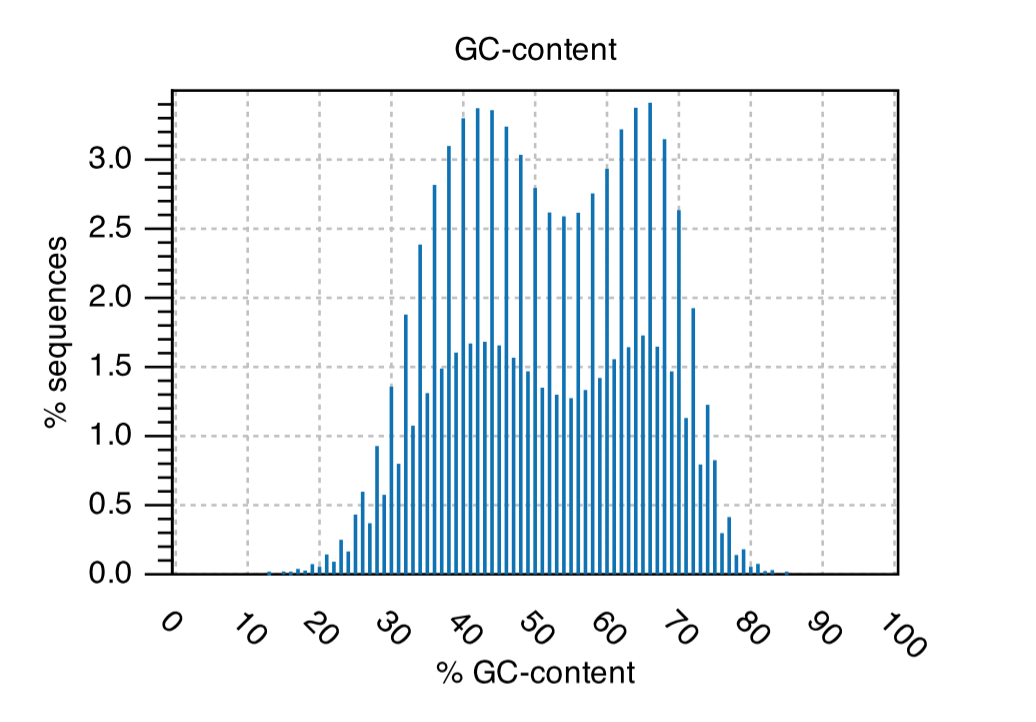
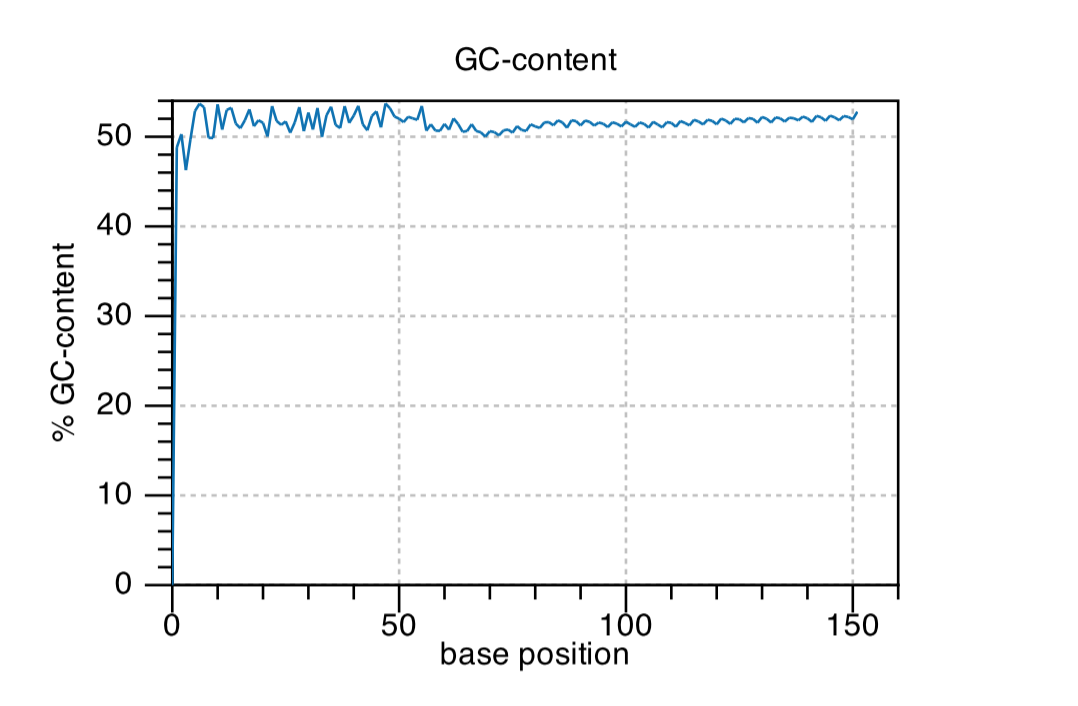
GC-content
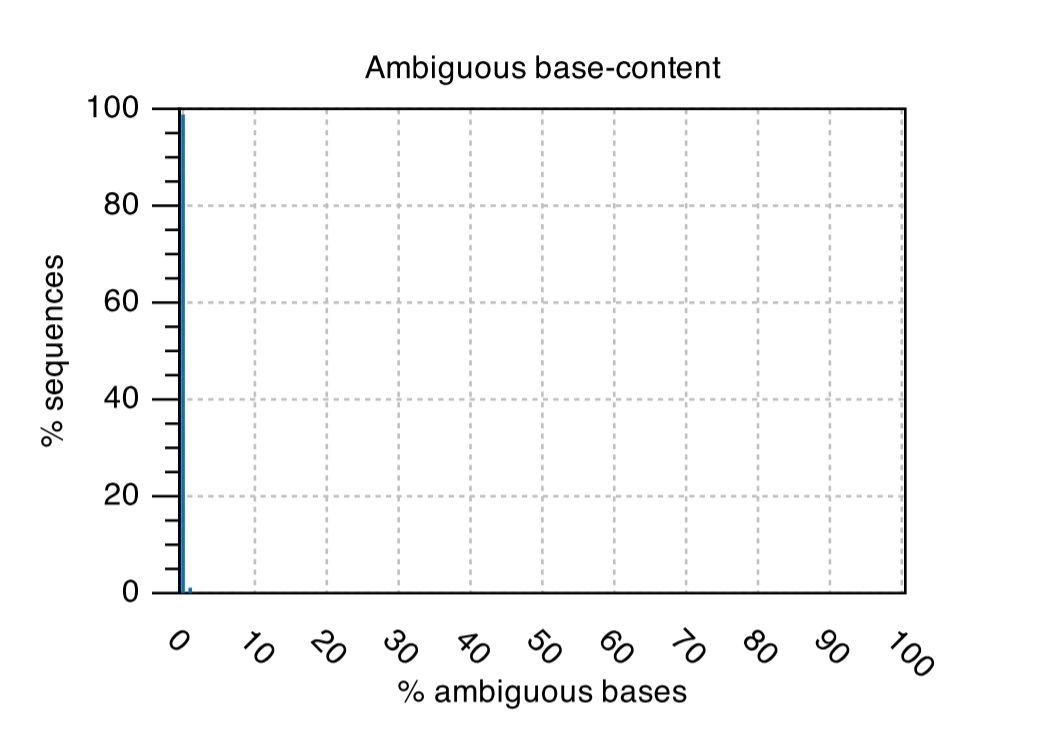
Ambiguous Base-content
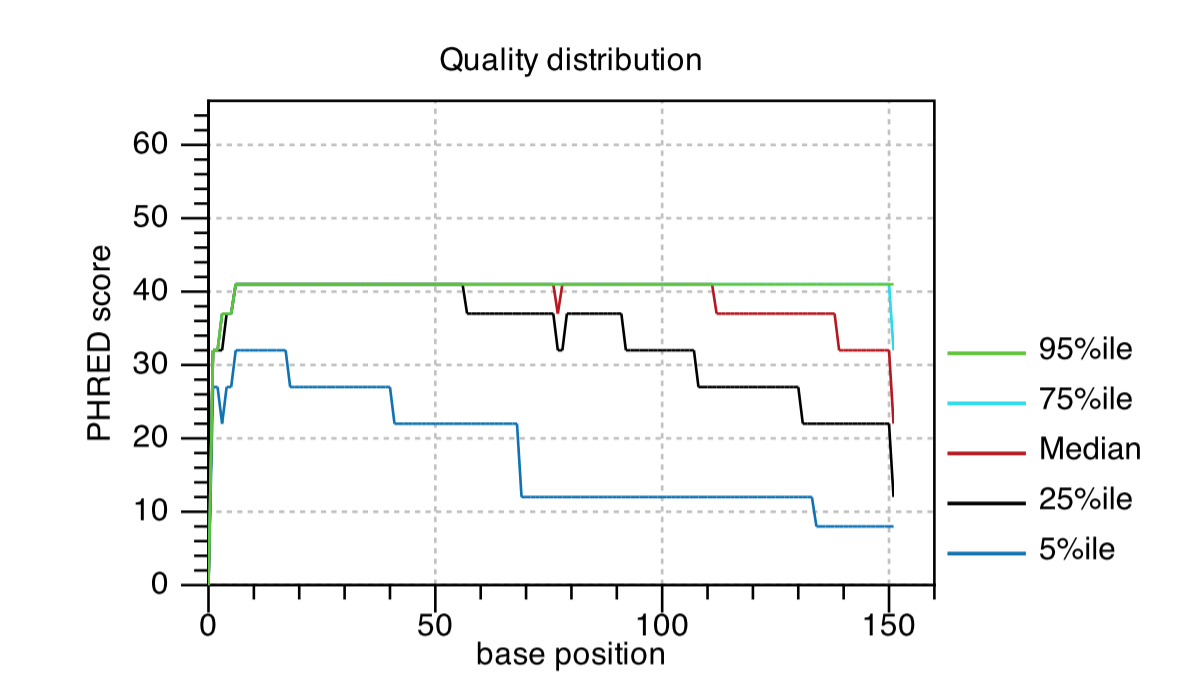
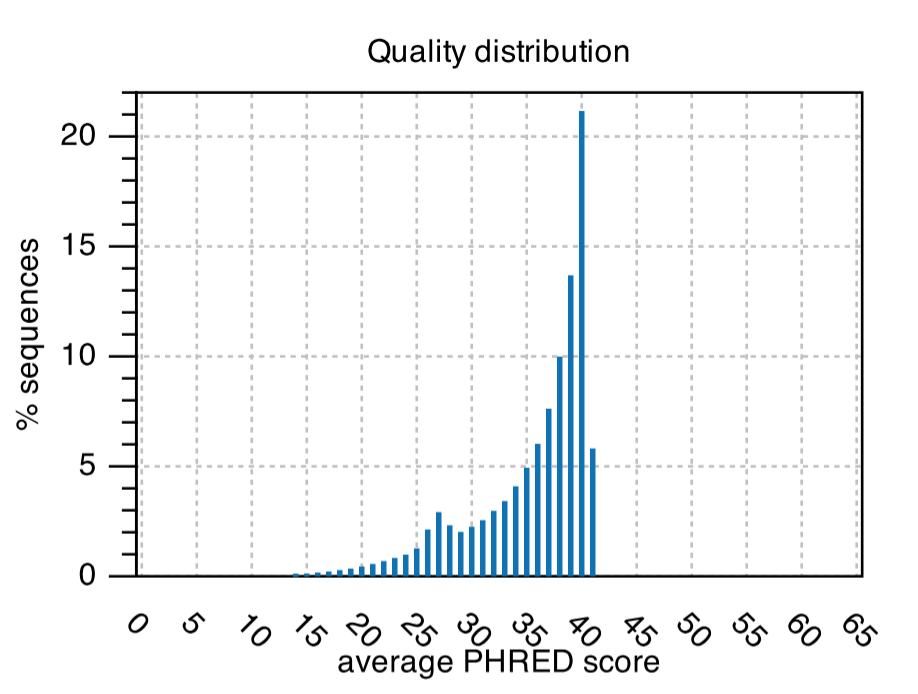
Quality Distribution
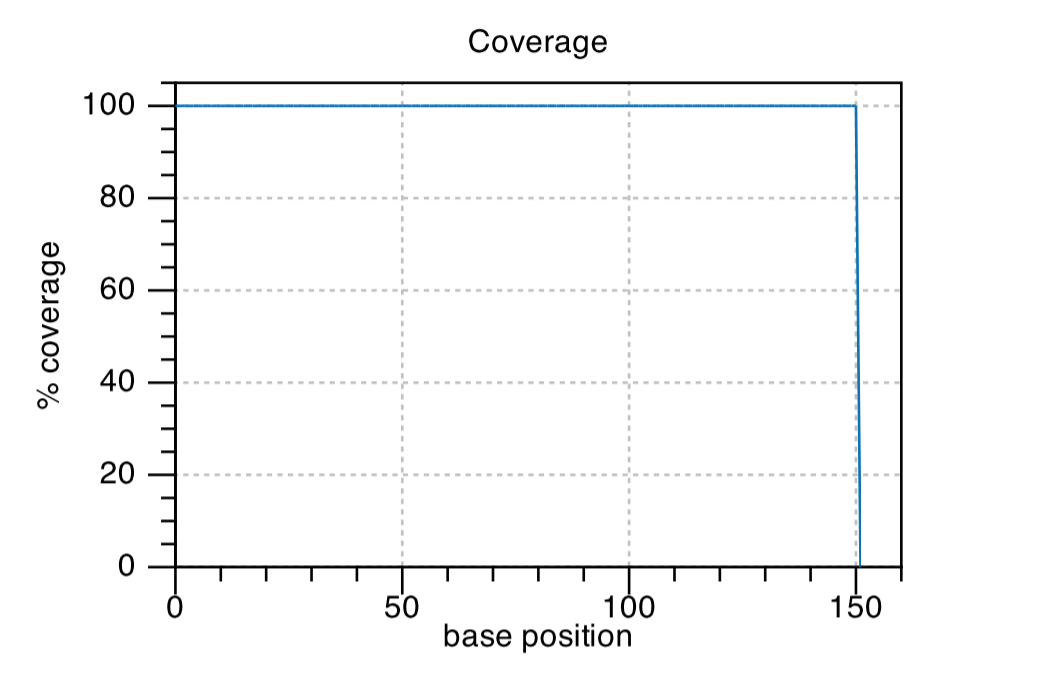
Coverage
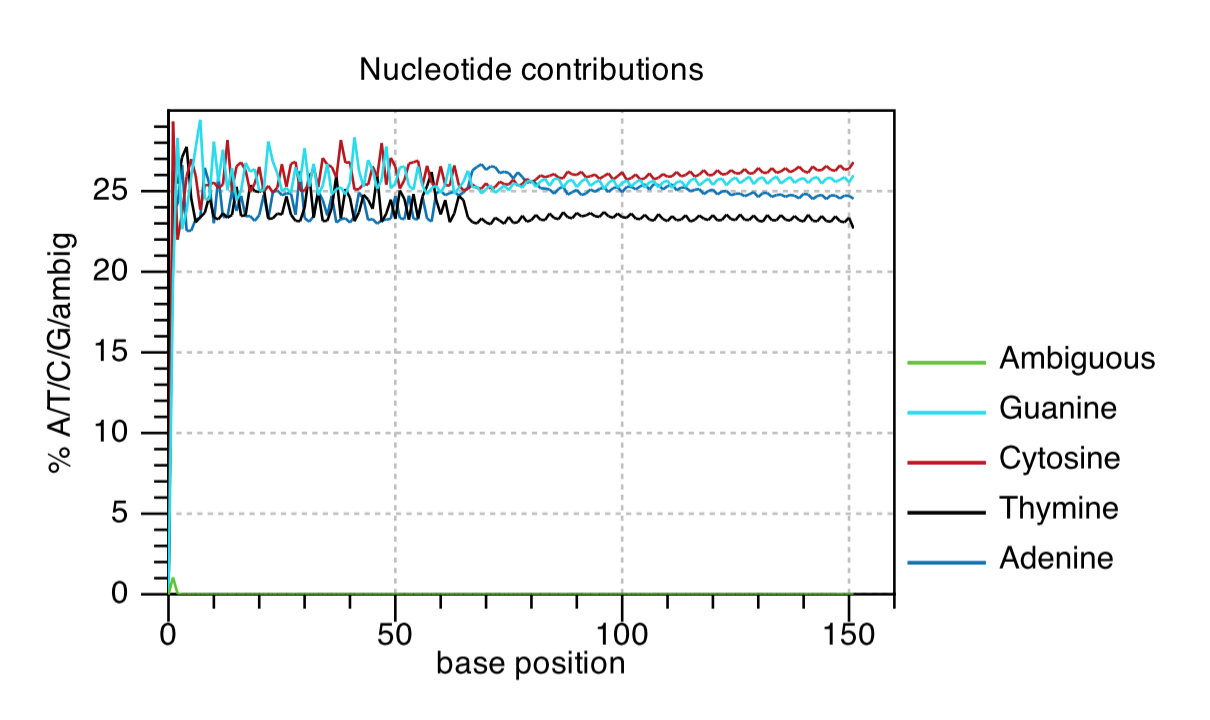
Nucleotide Contributions
Per-base GC Content
Quality Distribution