Quickstart
An overview of how to get started with CosmosID
Welcome to CosmosID! Here we will walk you through everything you need to get started with CosmosID, from creating an account to uploading samples and viewing results. If you would prefer to watch a video that takes you through the entire process, just click here.
In this guide we will go through the following steps:
 Follow the field prompts, review the terms of use, and check the box to accept. Click “Sign Up” and you will receive a welcome email.
If you already have an account and would like to sign in, enter your email address and password in the Sign In box. If you have forgotten your password, simply click “Forgot password?” in the Sign In box.
Navigate to the upload menu To expand the navigation menu, click the hamburger icon on upper left side of the screen:
Follow the field prompts, review the terms of use, and check the box to accept. Click “Sign Up” and you will receive a welcome email.
If you already have an account and would like to sign in, enter your email address and password in the Sign In box. If you have forgotten your password, simply click “Forgot password?” in the Sign In box.
Navigate to the upload menu To expand the navigation menu, click the hamburger icon on upper left side of the screen:
 Click “Upload” to navigate to the upload page.
Click “Upload” to navigate to the upload page.
 Once on the upload page, either drag and drop your files or click the page to open your file browser and select your samples that you would like to upload.
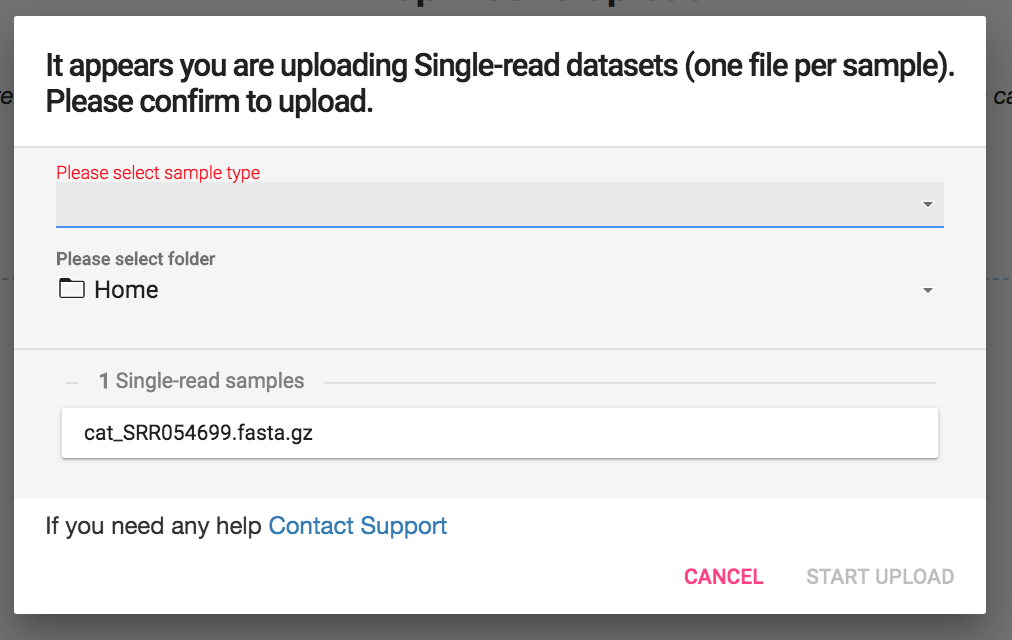
A box will open which will allow you to select your sample type:
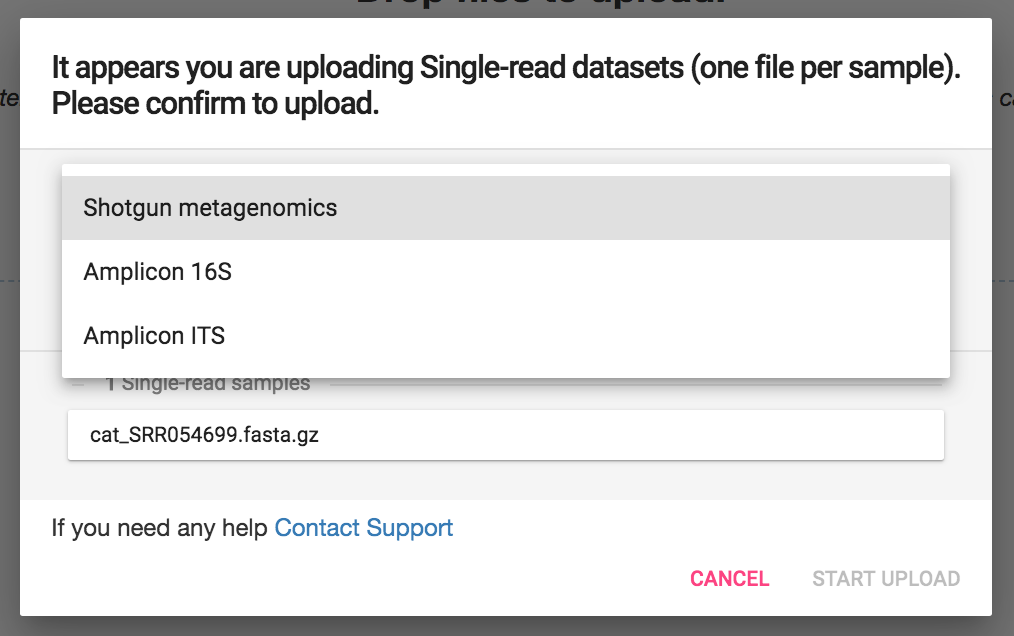
Select Shotgun Metagenomics, Amplicon 16S, or Amplicon ITS based on the type of analysis you will be performing.
Shotgun Metagenomics - using whole genome shotgun sequencing, the CosmosID algorithms identify microorganisms based on entire genomes represented in our database and in your samples
Amplicon 16S - the amplicon 16S is for bacterial 16S identification. Unlike shotgun metagenomics, amplicon 16S analysis looks only at the relevant bacterial 16S rRNA genes, not the entire genome for identification
Amplicon ITS - the amplicon ITS database is for fungal ITS identification.
Select your sample type:
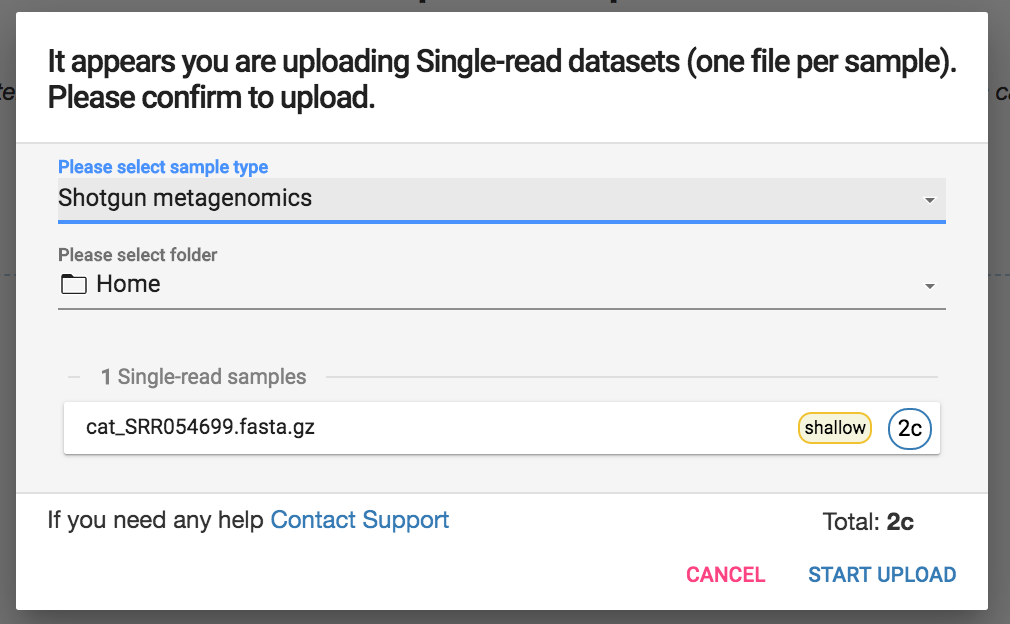
You also can select which folder in your account you would like the sample to be placed in.
Next to the sample name you will see the number of credits your account will be charged based on sample name and depth. Shotgun metagenomics samples with less than 0.5 GigaBases of sequence data are considered shallow sequencing samples and require 2 credits whereas those with more than 0.5 Gigabases of sequence data require 4 credits. Amplicon samples are 1 credit.
Click “Start Upload” to upload your samples.
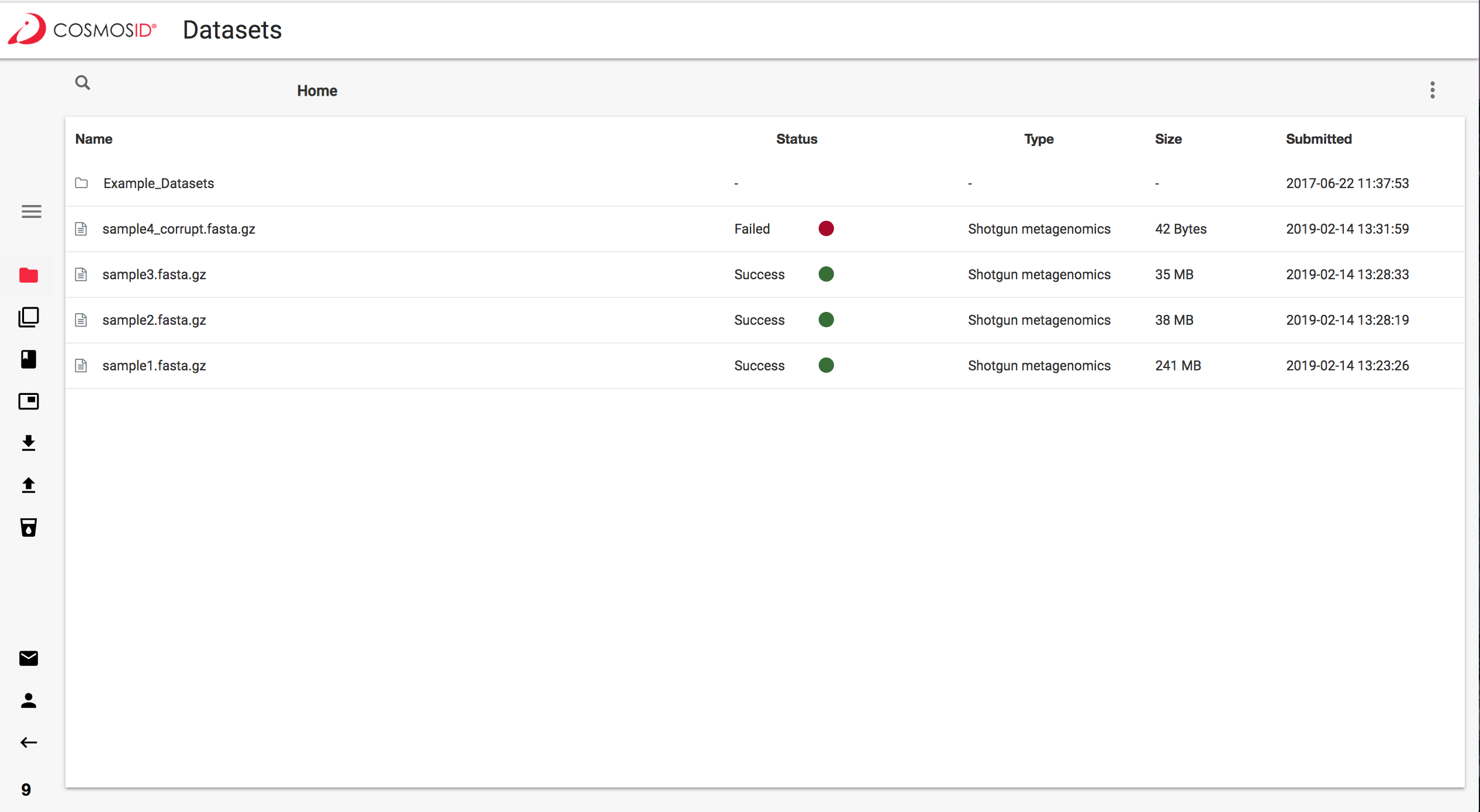
What a failed sample looks like:
Once on the upload page, either drag and drop your files or click the page to open your file browser and select your samples that you would like to upload.
A box will open which will allow you to select your sample type:
Select Shotgun Metagenomics, Amplicon 16S, or Amplicon ITS based on the type of analysis you will be performing.
Shotgun Metagenomics - using whole genome shotgun sequencing, the CosmosID algorithms identify microorganisms based on entire genomes represented in our database and in your samples
Amplicon 16S - the amplicon 16S is for bacterial 16S identification. Unlike shotgun metagenomics, amplicon 16S analysis looks only at the relevant bacterial 16S rRNA genes, not the entire genome for identification
Amplicon ITS - the amplicon ITS database is for fungal ITS identification.
Select your sample type:
You also can select which folder in your account you would like the sample to be placed in.
Next to the sample name you will see the number of credits your account will be charged based on sample name and depth. Shotgun metagenomics samples with less than 0.5 GigaBases of sequence data are considered shallow sequencing samples and require 2 credits whereas those with more than 0.5 Gigabases of sequence data require 4 credits. Amplicon samples are 1 credit.
Click “Start Upload” to upload your samples.
What a failed sample looks like:
 For details about samples, visualizations, filtering, changing taxonomic levels, etc., go to View Results.
All the details about the Example datasets can be found here: Definitions
For details about samples, visualizations, filtering, changing taxonomic levels, etc., go to View Results.
All the details about the Example datasets can be found here: Definitions
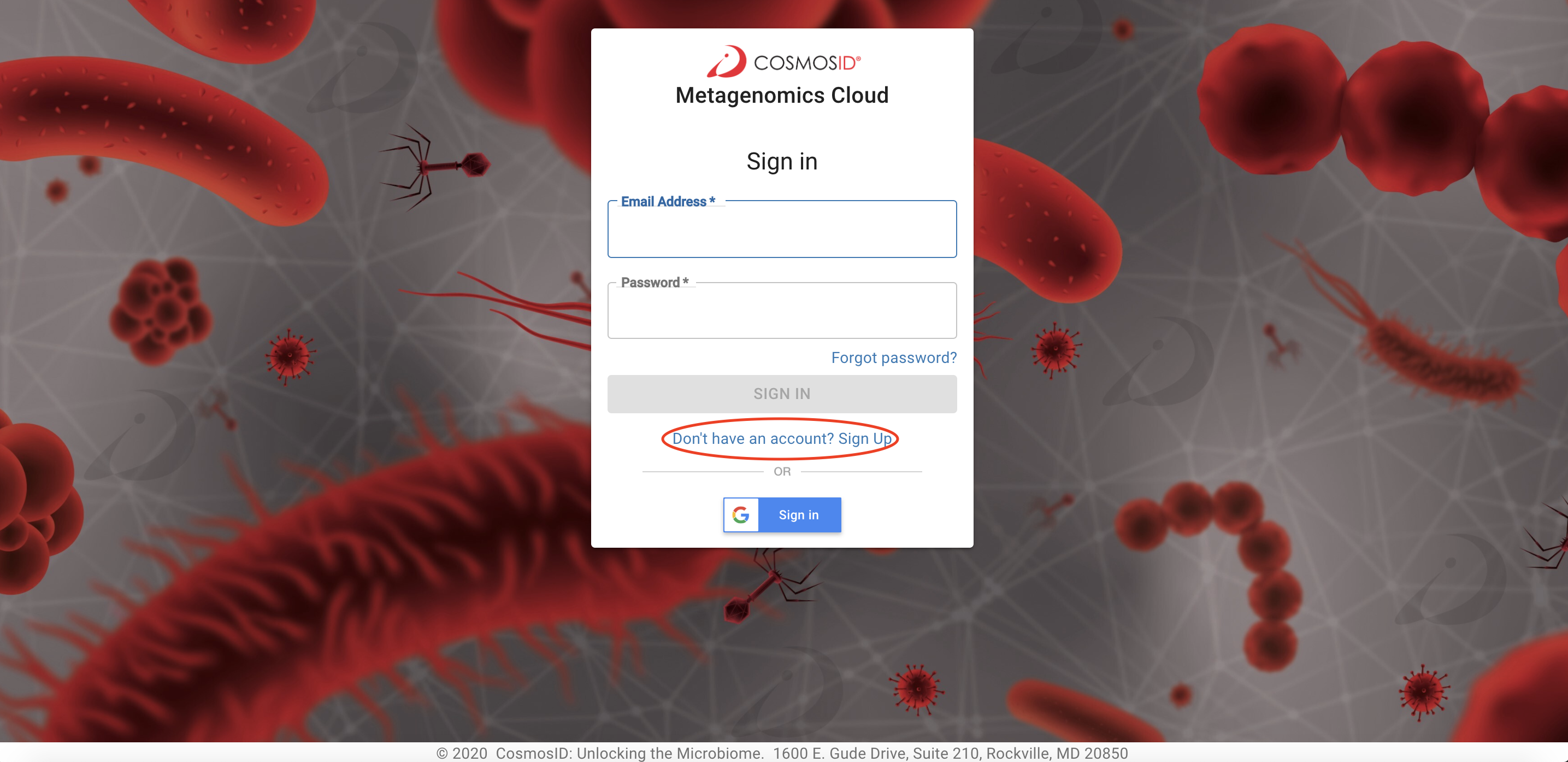
Step 1 Create An Account or Sign In
Navigate to the following page in your web browser https://app.cosmosid.com To create a new account, click “Sign Up” in the center of the log in page:
You also can sign in using gmailJust click the Google Sign In button!
Credits
If you would like to try out the app for free with your own samples, please email sales@cosmosid.comStep 2 Upload Your Samples
If you don’t have fasta/q files to upload, you can skip this step and use Example data.If you would like to load samples from BaseSpace, just click here.


Troubleshooting Sample Upload Failure
Occasionally a sample will fail to upload. Here are a number of possible causes:- Poor internet connection
- Internet connection interruption
- Browser closed while uploading
- If a sample is not in the proper format or fails to run for another reason, the status will show “failed” and you will not be charged a credit. Try to upload a correctly formatted file.
- If a sample upload has failed due to connectivity reasons, you can simply retry it.
- If you continue to experience problems with uploading or with a failed sample, please contact us on the app chat or support@cosmosid.com.

Step 3 Relax while your samples automatically run