Why Import SRA Data for Microbiome Analysis?Access to Publicly Available Data: Researchers can leverage publicly available SRA datasets to replicate studies, validate findings, or perform meta-analyses against their own data.Cost Efficiency: Using existing SRA datasets reduces the need for sequencing of new samples when performing secondary analyses or meta-analyses. By importing previously published data, users can rapidly evaluate different datasets or integrate multiple studies without the need to collect and sequence new samples.Extended Contextual Analysis: By analyzing previously published microbiome datasets from SRA, researchers can add context to their own data. This can provide valuable insights by comparing microbiomes across different populations, environments, diseases, or experimental conditions.Data Reuse for Novel Insights: Many research questions can be answered by revisiting older datasets with new hypotheses or more advanced analysis techniques. The Cosmos-Hub allows users to explore data from fresh perspectives using its comprehensive suite of microbiome analysis tools, potentially leading to novel discoveries.
Uploading SRA samples to the Cosmos-Hub
1
Navigate to the SRA Upload tool within the Upload Menu
Click “NCBI SRA” to navigate to the SRA import tool
2
Input your SRA IDs
SRA samples from Shotgun Metagenomic, 16S, or ITS samples can be input using SRR/DRR/ERR Accessions.Input up to 500 accessions (separated by comma, space, or semicolon) and click “Search”. The tool will find the data files (either single-end or paired-end) and report if any are unavailable.

3
Select your output folder and data type
NOTE: Folders must be pre-generated in the “Cohorts and Metadata” menu before uploading samples. You can also upload to the /Home folder and move the samples into subfolders after upload.You are required to select the TYPE of data from the following options:
- Shotgun Metagenomics - using whole genome shotgun sequencing, the Cosmos-Hub algorithms identify microorganisms based on entire genomes represented in our database and in your samples
- Amplicon 16S - the amplicon 16S is for bacterial 16S identification. Unlike shotgun metagenomics, amplicon 16S analysis looks only at the relevant bacterial 16S rRNA genes, not the entire genome for identification. This option requires the primer sequences used for generating libraries (e.g,. V1V2, V3V4, V4).
- You can select from standard primer sequences or input your own primer sequence
- Amplicon ITS - the amplicon ITS database is for fungal ITS identification.
4
Remove host reads
Selecting the sample host can remove host-reads for increased precision and accuracy in microbiome analysis, especially for functional profiling. You can select from various hosts, from humans to domestic cats!
If running CHAMP Human Profiler:Select “None” as the host when running the CHAMP Human Microbiome Profiler. CHAMP has a native host-removal algorithm for removing human host reads, and selecting a host for this pipeline will result in the sample analysis failing.
5
Upload your metadata (optional)
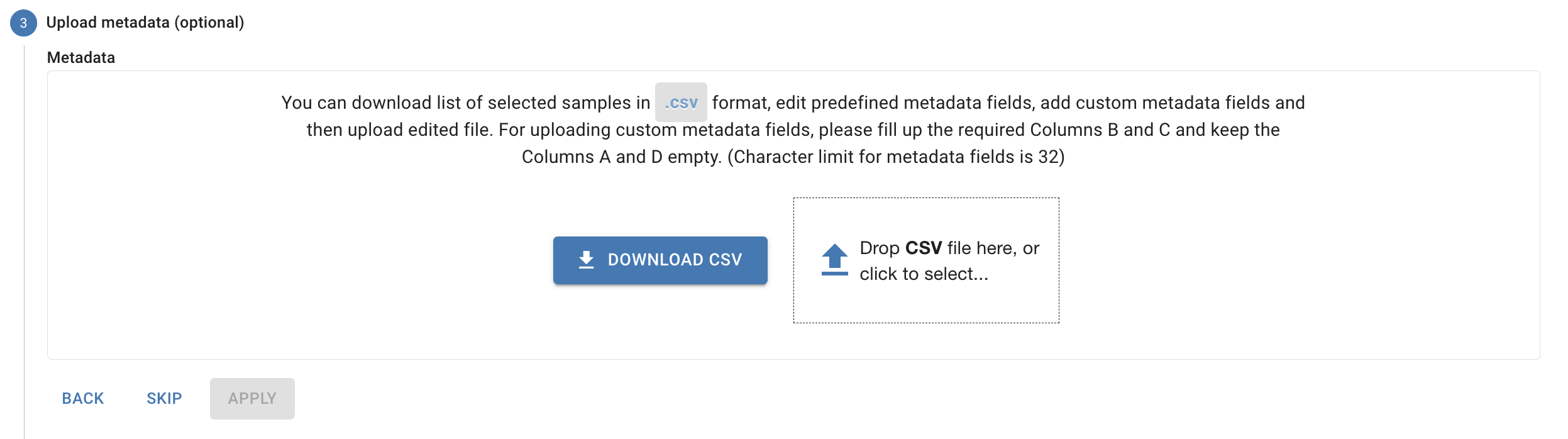
6
Select your workflow
7
Click "Start Upload"
| Taxa | Function | AMR and Virulence Marker | |
|---|---|---|---|
| WGS Deep | 4 | 4 | 2 |
| Amplicon 16S | 1 | N/A | N/A |
| Amplicon ITS | 1 | N/A | N/A |