What is Comparative Analysis?
Use comparative analysis to compare samples to one another using metadata-based cohorts. The analysis creates a matrix showing which organisms appear in each sample with useful visualizations and diversity figures.You must upload metadata or add tags before creating a comparative analysis.
To start, select your samples and click “Create CA”
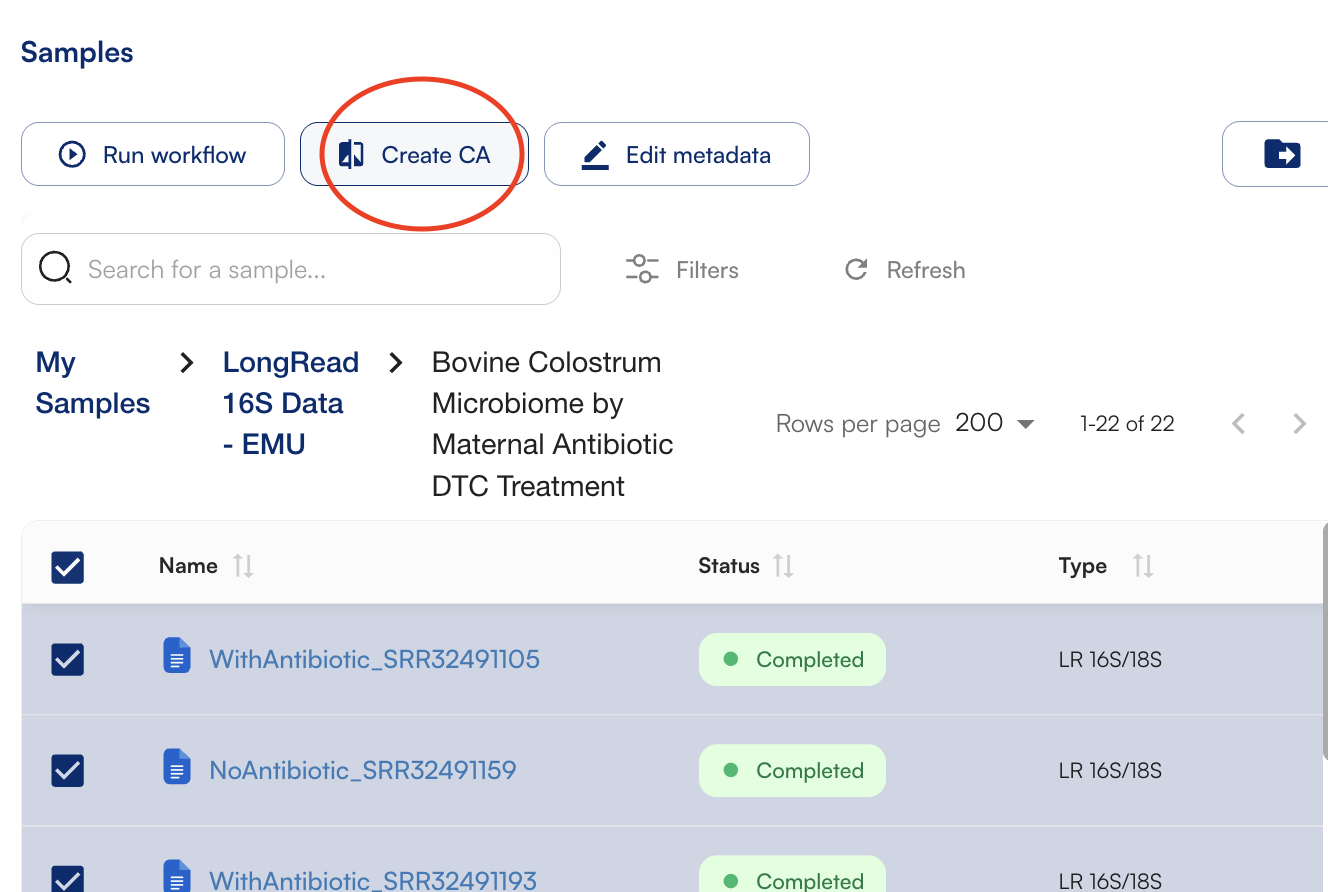
Group samples into cohorts based on metadata or tags
1. Using uploaded metadata
Give a descriptive name to your analysis file, and select the dropdown “Split cohorts by metadata” to view input metadata fields. For example, selecting the Antimicrobial Treatment variable, it will group samples into “Yes” and “No”. These variables can be any data you wish to input into the Hub. You can read how to upload metadata here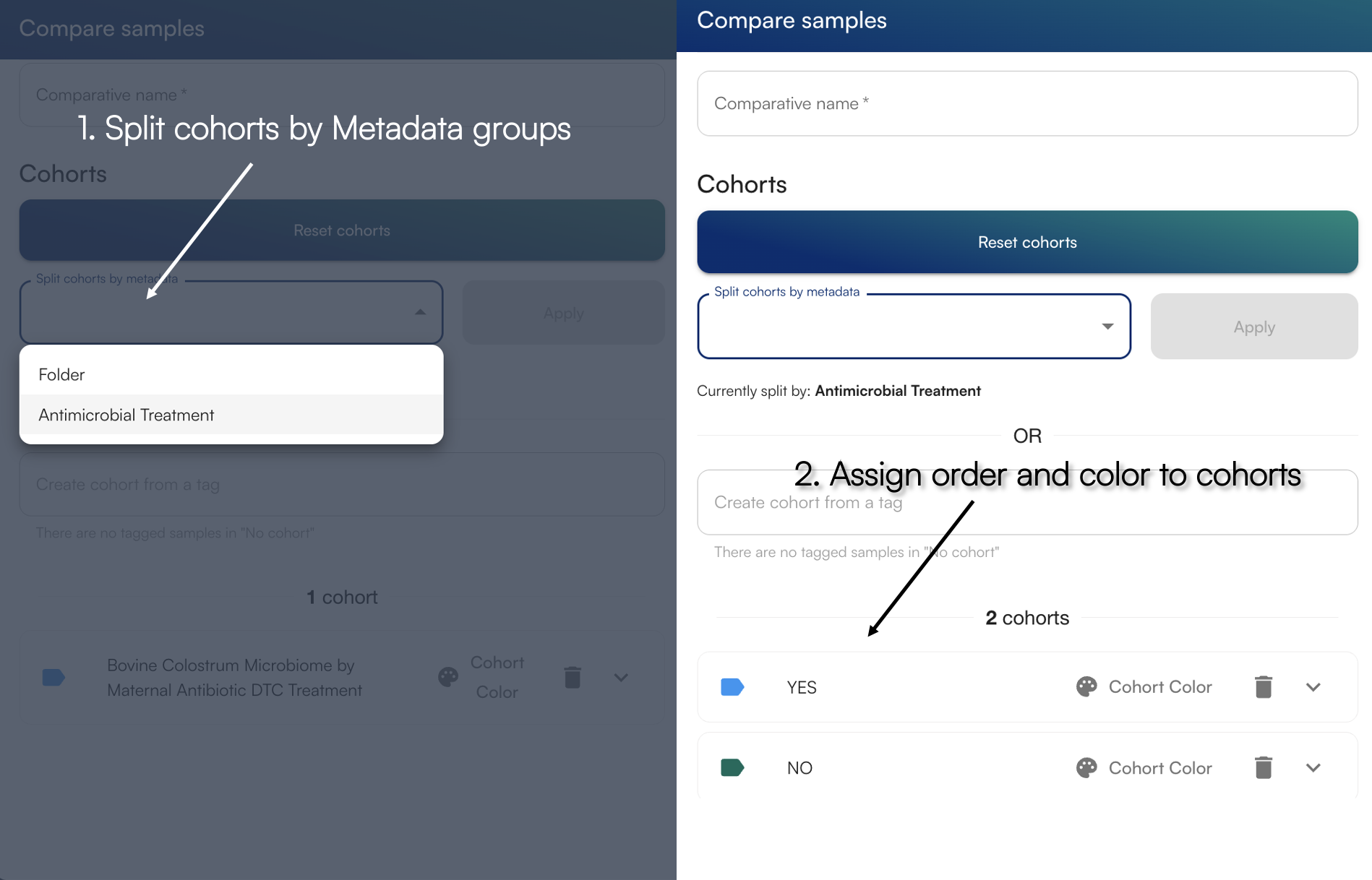
2. Using quick tags (#)
You can also define cohorts by adding custom tags to samples in the Samples menu, then use those tags to group samples.If you are still having trouble:
Watch the comprehensive tutorial here:Rename or delete analyses
- Click the comparative analysis icon in the left panel
- Select one or more analyses
- Click the Delete icon (you can delete multiple at once)
- Confirm deletion when prompted
- Note: Deleted analyses are permanently removed but can be recreated from original files
- Select only one analysis
- Click the Rename icon
- Enter a new name using allowed characters: letters, numbers,
',&,(,),[,],+,!,.,-,_