Read the LEfSe paper here: Segata, N., Izard, J., Waldron, L. et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60 (2011). https://doi.org/10.1186/gb-2011-12-6-r60
What is LEfSe Biomarker Discovery Analysis?
LEfSe (Linear discriminant analysis effect size) is an algorithm for High-Dimensional biomarker discovery that identifies genomic features (genes, pathways, or taxa) characterizing the differences between two or more biological conditions. It emphasizes both statistical significance and biological relevance, allowing researchers to identify discriminative features that are that are statistically different among biological classes. Specifically, the non-parametric factorial Kruskal-Wallis (KW) sum-rank test is used to detect features with significant differential abundance with respect to the class of interest. As a last step, LEfSe uses Linear Discriminant Analysis to estimate the effect size of each differentially abundant feature and rank the feature accordingly.How do you generate LEfSe biomarker discovery analysis?
In oder to run LEfSe biomarker discovery analysis, at-least 2 datasets and 2 cohorts are required for comparative analysis creation. 2 parameters are required for LEFSE biomarker discovery analysis.- Alpha: Alpha is a threshold value for non parametric factorial Kruskal-Wallis test which is used to judge whether a test statistic is statistically significant when comparing two or more cohorts or groups
- Threshold: Threshold represents the linear discriminant analysis score or effect size cutoff value for discriminative biomarker or feature.
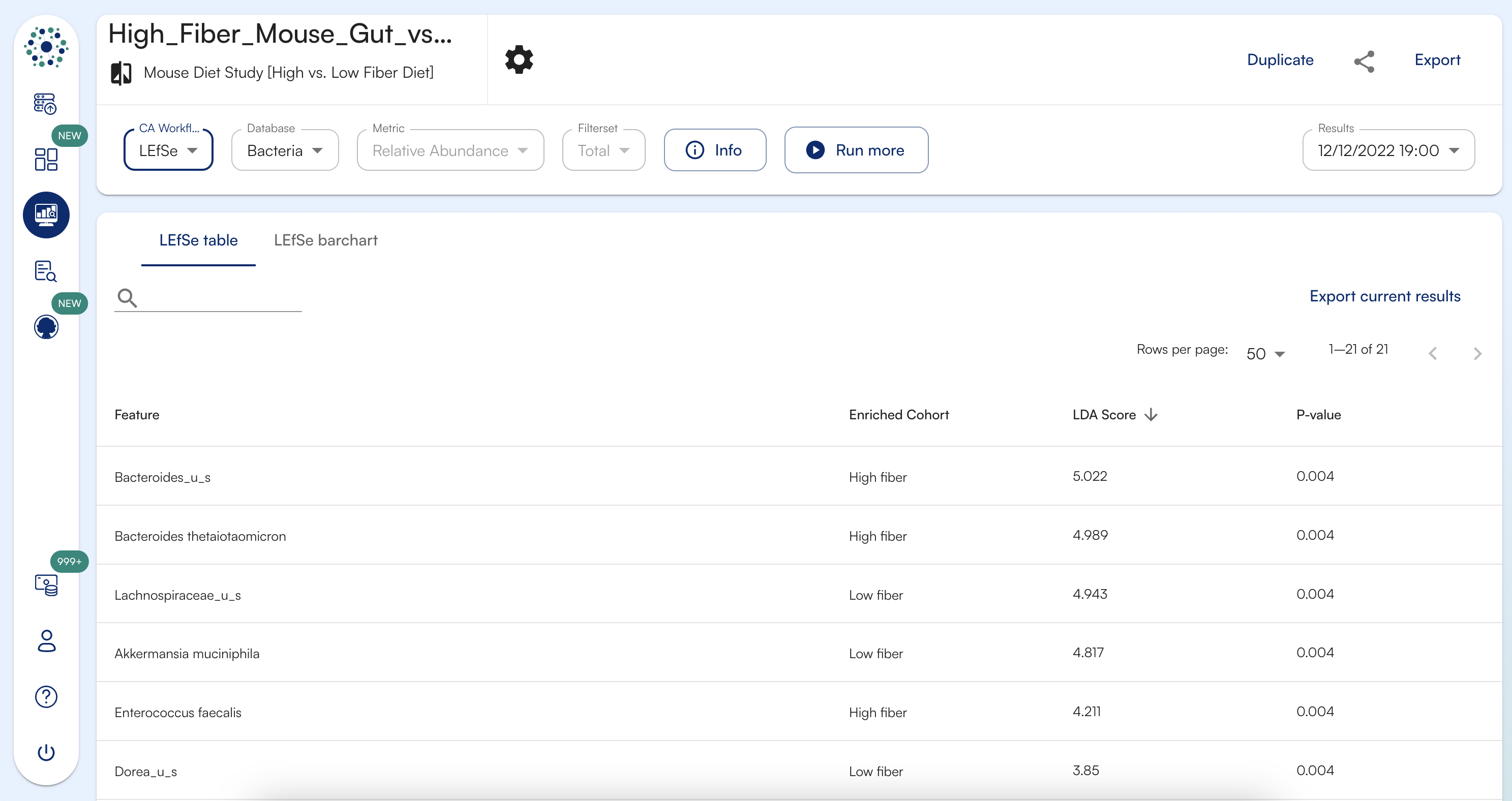
What do the columns represent in LEfSe table results view?
LEFSE Table comprises of 4 columns. The column description is given below.- Feature: The feature column lists out the feature name that has been found to be discriminative among cohorts
- Enriched Cohort: The enriched column lists out the cohort name in which the respective feature has been found to be discriminative
- LDA: LDA represents the Linear Discriminant Analysis score or effect size for discriminative biomarker or feature
- P-value: P-value represents the p-value for factorial Kruskal-Wallis test among cohort
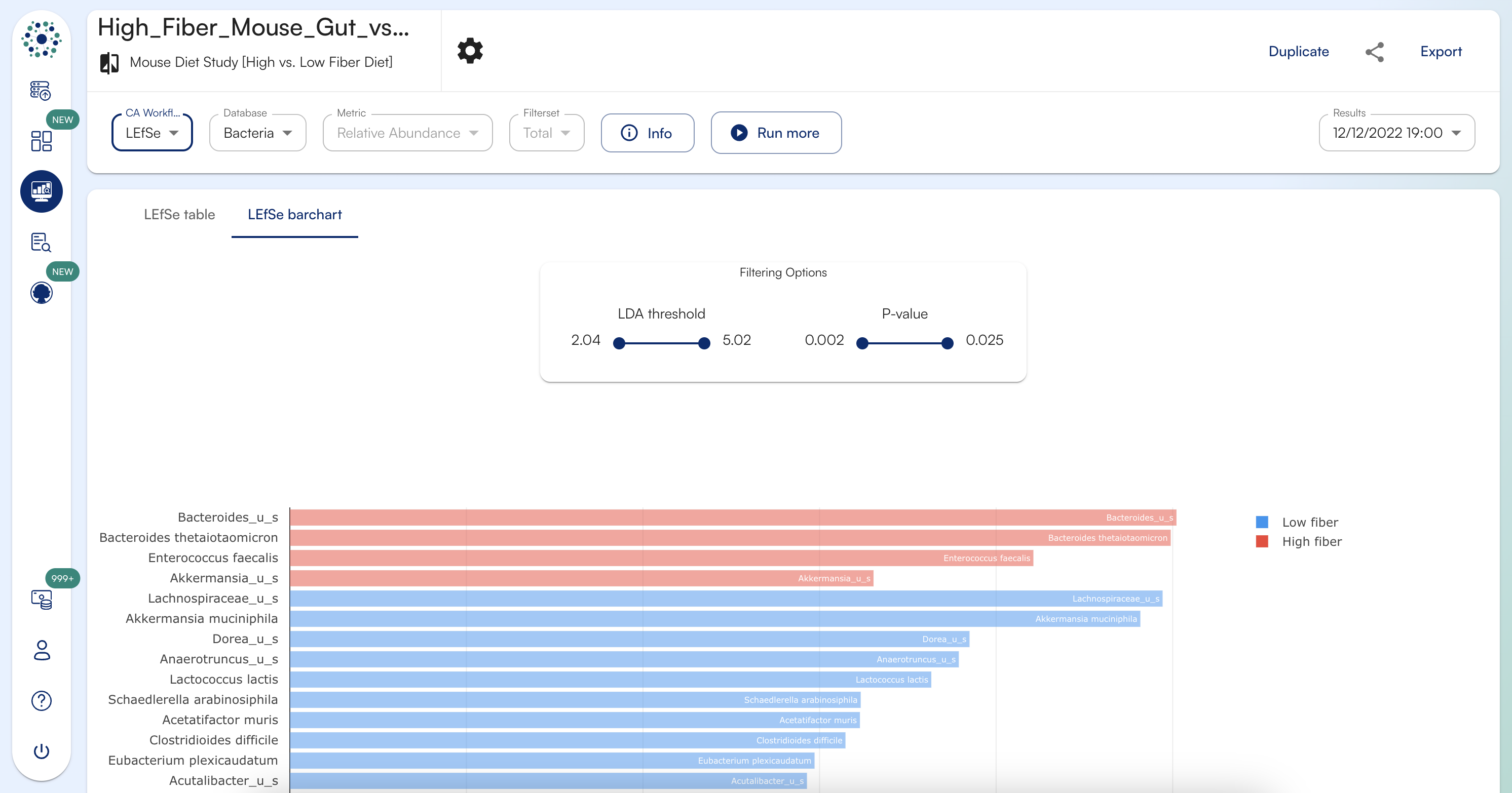
How do you interpret LEfSe Barchart results view?
LEfSe Barchart is a visual representation of discriminative features/biomarkers found by LEfSe tool ranking them accordingly to their LDA Score/Effect Size. The barchart is dynamic and can be filtered using both LDA score and P-value.