There are 4 upload methods to choose from:
Upload from your Computer
Upload local FASTQ, SRA, or BAM files via drag-and-drop.
Link a BaseSpace Account
Connect your Illumina BaseSpace account and import data directly.
Import from NCBI SRA
Use public SRA accession numbers to fetch data.
Use the Command Line Interface
Use our CLI or API for large batch uploads and automation.(Click for API documentation)
Select Your Data Type
- Amplicon (16S / ITS)
- Shotgun Metagenomics
- SRA Upload
Amplicon pipelines are used to identify bacterial or fungal communities based on specific genes. These workflows support both single-end and paired-end reads.
Requirements
Requirements
- Read type: Paired-end or single-end
- Min samples: 10 (for ASV/DADA2 workflows)
- File size: ≤ 1 GB uncompressed (250 MB compressed)
- PHRED score ≥ 20
-
Formats:
.fastq.gz,.fq.gz,.fasta.gz,.sra.gz -
Paired-end naming:
SampleID_R1_001.fastq.gzSampleID_R2_001.fastq.gz
- Reads must overlap ≥ 15 bp
-
Watch the 16S Tutorial Video
Workflow Options
Workflow Options
- ASV 16S rRNA Profiling (DADA2-based)
- OTU ITS rRNA Profiling (QIIME2-based)
Common Pitfals
Common Pitfals
- Filenames with spaces or special characters will break pairing
- Unpaired files will appear as “Single-read samples”
Uploading Metadata
You can upload a metadata CSV during or after your data upload through the Cohorts and Metadata page.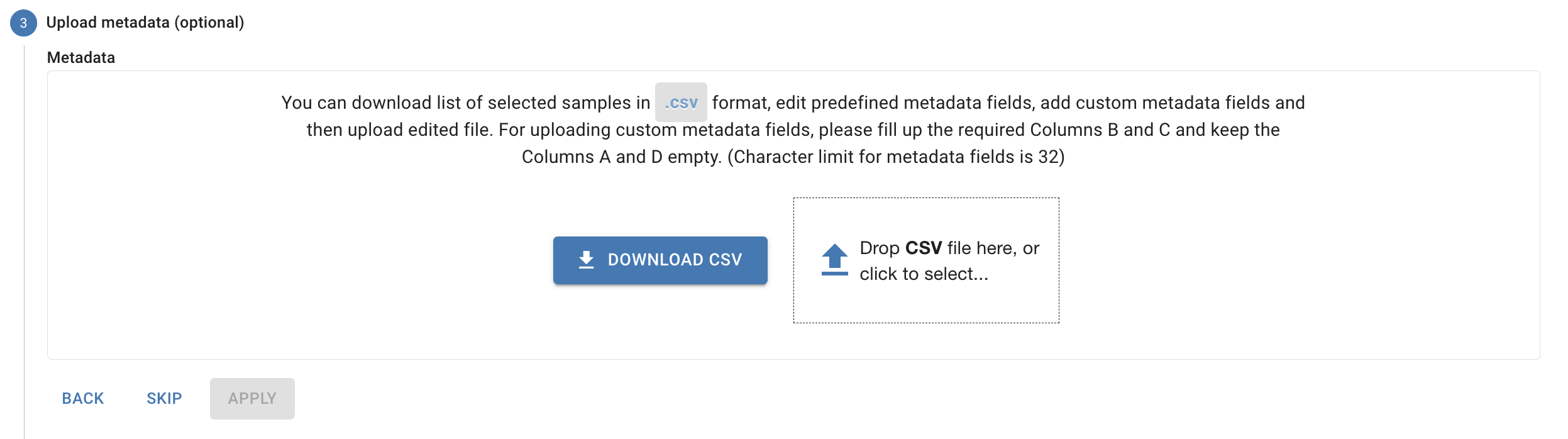
Workflow Selection
After upload, choose a workflow that matches your data type. The Cosmos-Hub will only show valid options based on your selections.
Other notes:
- If a sample fails to upload, you can retry it
- If a sample is not in the proper format or fails to run for another reason, the status will show “failed” and you will not be charged credits.
- If you have problems uploading or with a failed sample, please submit a help ticket or email us at help@cosmos-hub.com.